How To Read Microarray Heat Map
In these arrays mRNA is copied into cDNA and then labeled and hybridized to an array that has all human or mouse or rat etc. In heat maps the data is displayed in a grid where each row represents a gene and each column represents a sample.
 Heat Map Of Microrna Mirna Microarray Expression Data From Plasma Samples Of Patients With Ventricular Septal Defect Vsd N 3 And Vsd Free Controls N 3
Heat Map Of Microrna Mirna Microarray Expression Data From Plasma Samples Of Patients With Ventricular Septal Defect Vsd N 3 And Vsd Free Controls N 3
The basic idea of a heat map is that the graph is divided into rectangles or squares each representing one cell on the data table one row and one data set.
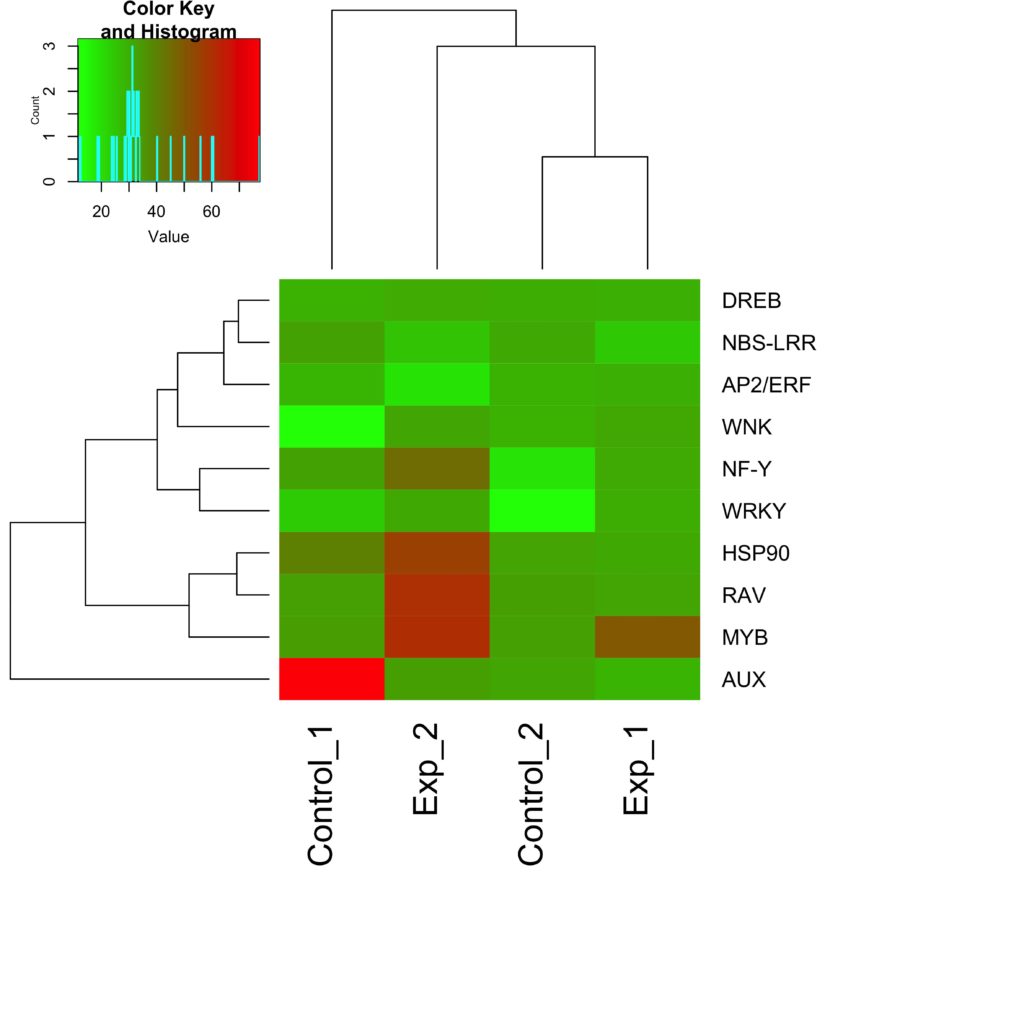
How to read microarray heat map. Thus according to your heat map you have values ranging from -20 to 20 and you can see from the histogram that you have more positive than negative values. The first section of this page uses R to analyse an Acute lymphocytic leukemia ALL microarray dataset producing a heatmap with dendrograms of genes differentially expressed between two types of leukemia. Start a free app analysis.
Heatmapper is a versatile tool that allows users to easily create a wide variety of heat maps for many different data types and applications. Each row of the heat map represents a probe. The colour and intensity of the boxes is used to represent changes not absolute values of gene expression.
The colors of the heat map are normalized expression values. Im guessing that since your heat map scale is -20 to 20 more or less centered at 0 you are actually plotting log ratios and perhaps dont realize it. Each column is a variable.
Have RNA-seq or microarray data. Color coding after computing z-scores row scaling This looks much better and you can see patterns picked out by the clustering algorithm. I have hinted in Part 1 of this series that gene expression profiling using microarrays is a prime application for heatmaps.
Try our mobile app heatmaps for free to get started with the analysis. Heatmapper is a freely available web server that allows users to interactively visualize their data in the form of heat maps through an easy-to-use graphical interface. A heat map is a well-received approach to illustrate gene expression data.
If you work in any area of quantitative biology and especially if you work with transcriptomic data then you are probably familiar with heatmaps used for as long as I have been in research these figures cluster rows and columns of a data matrix and show both dendrograms alongside a colour-scaled representation of the data matrix itself. The most commonly used one is an expression microarray. A heat map or heatmap is a data visualization technique that shows magnitude of a phenomenon as color in two dimensions.
Today we will look at the differences of gene expression in Acute Lymphoblastic Leukemia ALL samples that have either no cytogenetic abnormalities or the famous BCRABL chromosomal translocation Philadelphia chromosome. Prism offers lots of options to make Heat maps. There is a follow on page dealing with how to do this from Python using RPy.
Microarray heatmap 64k views. The trick is that usually 2 different cell types are used for example a healthy cell and a cancer cell. Microarray Data Analysis M.
How to Read a Heatmap. You can transpose the matrix with tdata to swap X and Y axis. It is time to deal with some real data.
Generates color-coded Clustered Image Maps CIMs heat maps to represent high-dimensional data sets such as gene expression profiles. Interpreting Heat Map Visualizations. Heat maps are a standard way to plot grouped data.
Learn how to interpret data presented in heat map visualizations. Each observation is a row. Madan Babu Abstract This chapter aims to provide an introduction to the analysis of gene expression data obtained using microarray experiments.
The cluster heat map and the spatial heat map. The variation in color may be by hue or intensity giving obvious visual cues to the reader about how the phenomenon is clustered or varies over spaceThere are two fundamentally different categories of heat maps. How to read a heat map is a craft.
In the example below red represents up-regulated genes and blue represents down-regulated genes. It has been divided into four sections. What possible tools can help you find your genes of interest.
The original citation for the raw data is Gene expression profile of adult T-cell acute lymphocytic. Consider heat map visualization configuration options. Please note that all of the default for colors on most of these heat maps is red and green.
A snapshot of some of the data that can be exported from an eye tracking experiment. And here is the correlation distance heat map after converting to z-scores of the rows genes. Each square is a value the closer to yellow the higher.
With this list you are well prepared for your next analysis. The heat map makes for an accessible and understandable framing of the data but if you want to know more about what underlies it then youll need to export the numbers for further analysis. The fi rst section provides basic concepts on the working of microarrays and describes the basic principles behind a microarray.
We introduced CIMs in the mid-1990s for data on drug activity target expression gene expression and proteomic profiles. Is there any pattern in your expression data. 5 Things You Have to Know.
How to read it. It is an impressive visual exhibit that addresses explosive amounts of NGS data. To read you data into R see readtable assuming it is in a csv format.
I want to have a heat map showing the expression pattern of my interest probsets in this array for example in this array i have 4 varieties and different tissues rind and flesh and phases 010203040 and 50 days after. The heatmap is a visualization of the microarray values for the returned probes of interest. I know you are totally at sea but heat maps are now commonly used to help.
Each column of the heat map either represents a tissue sample or anatomical brain structure depending on the selected resolution see below. The rectangle or square is color coded according to the value of that cell in the table.
 A Heatmap Showing Differential Mirna Expression In Diseased Vs Normal Download Scientific Diagram
A Heatmap Showing Differential Mirna Expression In Diseased Vs Normal Download Scientific Diagram
 Biological Interpretation Of Gene Expression Data Functional Genomics Ii
Biological Interpretation Of Gene Expression Data Functional Genomics Ii
 How Can I Generate A Heatmap And Clustering Of Differentially Expressed Genes In A Rna Seq Data
How Can I Generate A Heatmap And Clustering Of Differentially Expressed Genes In A Rna Seq Data
 Heat Map Displaying Hierarchical Clustering Results From Microarray Expression Data Of Genes Implicated In The Toll Like Receptor Signalling Pathway
Heat Map Displaying Hierarchical Clustering Results From Microarray Expression Data Of Genes Implicated In The Toll Like Receptor Signalling Pathway
 Heat Map Representing The Log2 Fold Change Values Between Contrasted Treatments For The 844 Genes Found To Be Differentially Expressed In At Least One Of The Contrasts
Heat Map Representing The Log2 Fold Change Values Between Contrasted Treatments For The 844 Genes Found To Be Differentially Expressed In At Least One Of The Contrasts
 How To Plot Gene Expression Heatmap Based On Groups In R
How To Plot Gene Expression Heatmap Based On Groups In R
 Pdf The History Of The Cluster Heat Map
Pdf The History Of The Cluster Heat Map
 Jci Spdef Is Required For Mouse Pulmonary Goblet Cell Differentiation And Regulates A Network Of Genes Associated With Mucus Production
Jci Spdef Is Required For Mouse Pulmonary Goblet Cell Differentiation And Regulates A Network Of Genes Associated With Mucus Production
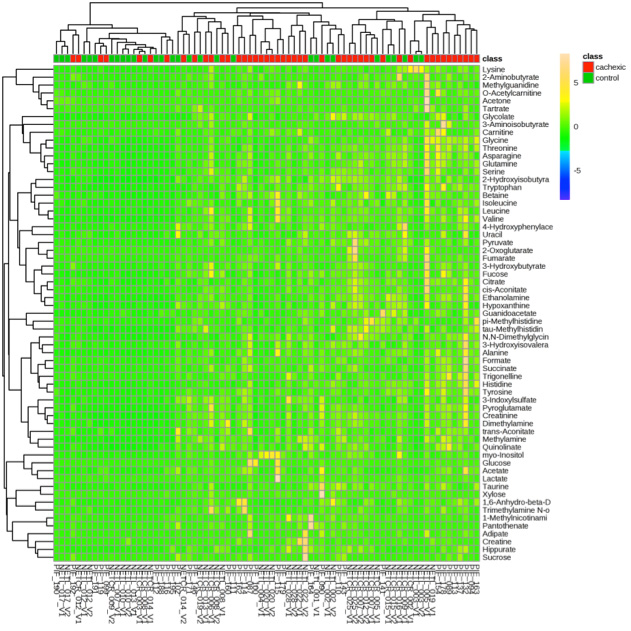 Heatmap Service Creative Proteomics
Heatmap Service Creative Proteomics
You Probably Don T Understand Heatmaps
When Using Heatmap 2 From R To Make A Heatmap Of Microarray Data How Are The Genes Clustered
 Heatmap Based On The Results Of Sam One Class Analysis Of The Microarray Data From 6 Thyroid Cancer Cell Lines And Compared To Tumor Tissues
Heatmap Based On The Results Of Sam One Class Analysis Of The Microarray Data From 6 Thyroid Cancer Cell Lines And Compared To Tumor Tissues
 Sigma Aldrich Advanced Genomics Archives Page 3 Of 10 Bitesize Bio
Sigma Aldrich Advanced Genomics Archives Page 3 Of 10 Bitesize Bio
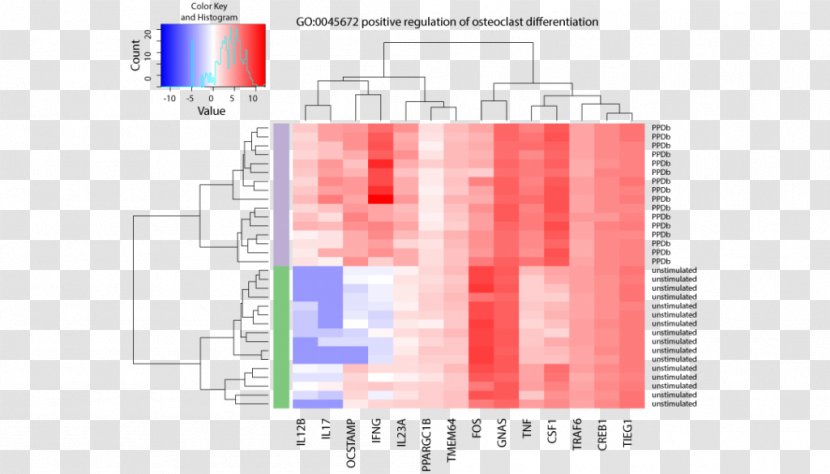 Dna Microarray Heat Map Rna Seq Nucleic Acid Sequence Rnaseq Gene Expression Transparent Png
Dna Microarray Heat Map Rna Seq Nucleic Acid Sequence Rnaseq Gene Expression Transparent Png
 How Can We Covert A Heatmap Of Two Channel Microarray Data
How Can We Covert A Heatmap Of Two Channel Microarray Data
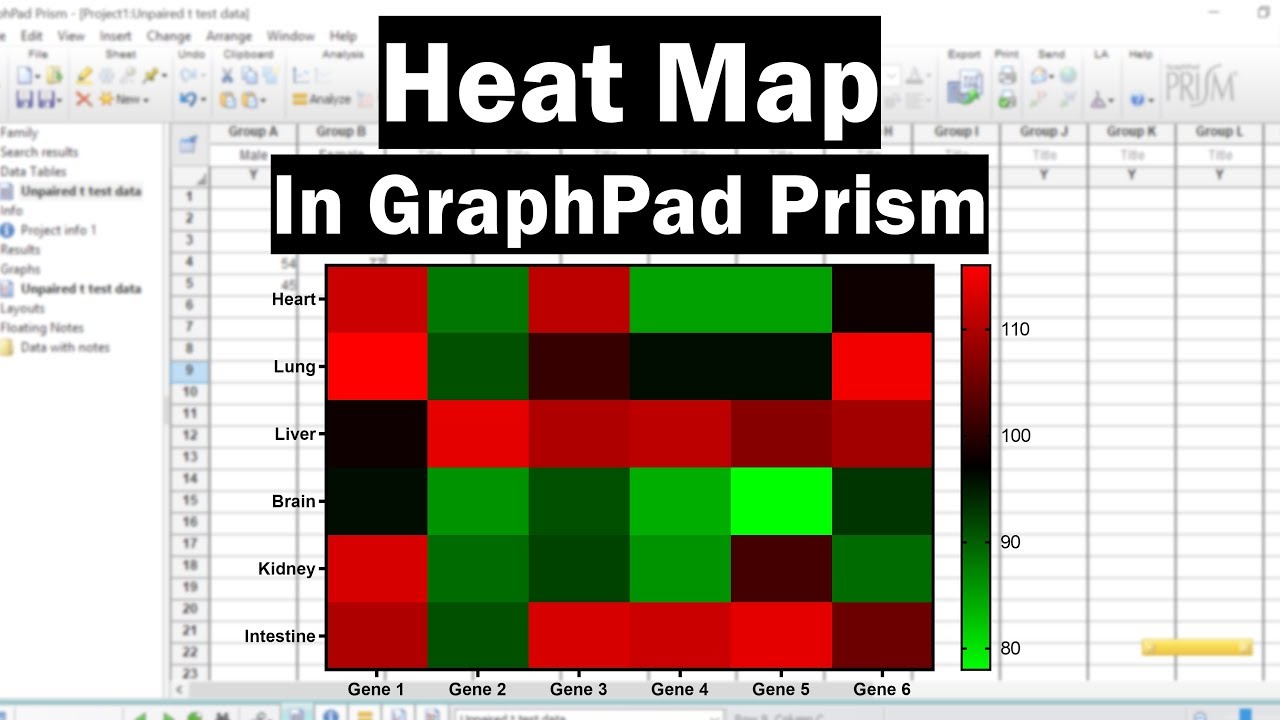 How To Create A Heat Map In Graphpad Prism Youtube
How To Create A Heat Map In Graphpad Prism Youtube
 Heat Map Of The Microarray Gene Expression Profile Of Hepg2 Cells Treated With Live Dengue Virus Tsv01 V Or Heat Inactivated Tsv01 H Transcripts Identified As Being Differentially Expressed By Sam Analysis
Heat Map Of The Microarray Gene Expression Profile Of Hepg2 Cells Treated With Live Dengue Virus Tsv01 V Or Heat Inactivated Tsv01 H Transcripts Identified As Being Differentially Expressed By Sam Analysis

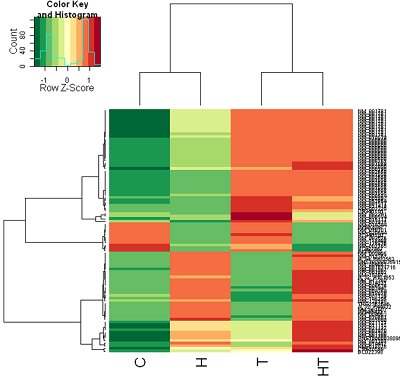
Post a Comment for "How To Read Microarray Heat Map"